Metabolic Fingerprinting
Metabolic fingerprinting refers to the use of machine output as potentially recognizable chemical pattern, specific of an individual sample. Metabolite fingerprinting by NMR is a fast, convenient, and effective tool for discriminating between groups of related samples and it identifies the most important regions of the spectra for further analysis.
So the spectra processing is an intermediate step between raw spectra and data analysis. It consists to preserve as much as possible the variance relative to the chemical compounds contained in the NMR spectra while reducing other types of variance induced by different sources of bias such as baseline and misalignment. See Spectra processing section.
Then, the identity of the metabolites of interest is established after statistical data analysis of metabolic fingerprints, and this involves to be able:
- to highlight that spectral regions having a difference between the groups are statistically significant.
- to ensure that each of these regions involves only a single metabolite, i.e. there is unique correspondence between a bucket and a resonance (spectral signature) of a metabolite
The standard approach in NMR-based metabolomics implies the division of spectra into equally sized bins, thereby simplifying subsequent data analysis. Yet, disadvantages are the loss of information and the occurrence of artifacts caused by peak shifts. Therefore we implemented the Adaptive Intelligent Binning (AI-Binning) algorithm which largely circumvents these problems. It recursively identifies bin edges in existing bins, requires only minimal user input, and avoids the use of arbitrary parameters or reference spectra. This algorithm is well adapted to meet the second point mentioned above.
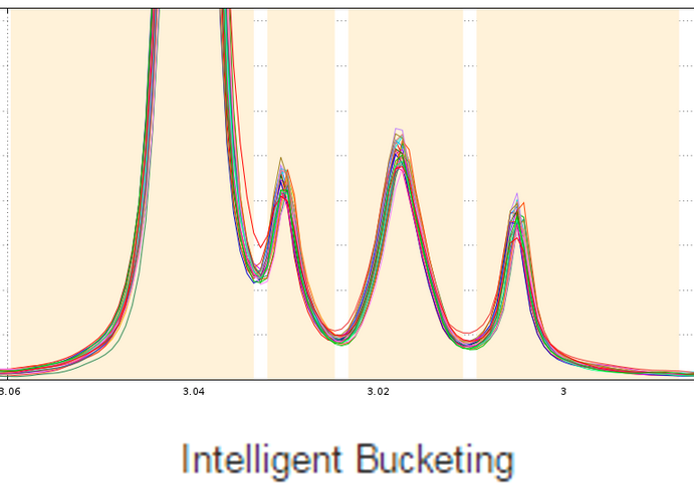
De Meyer T., Sinnaeve D., Gasse B., Tsiporkova E., Rietzschel E., De Buyzere M., Gillebert T., Bekaert S., Martins J. and Criekinge W. (2008) NMR-Based Characterization of Metabolic Alterations in Hypertension Using an Adaptive, Intelligent Binning Algorithm. Analytical Chemistry 80(10):3783–3790
How to further proceed ?
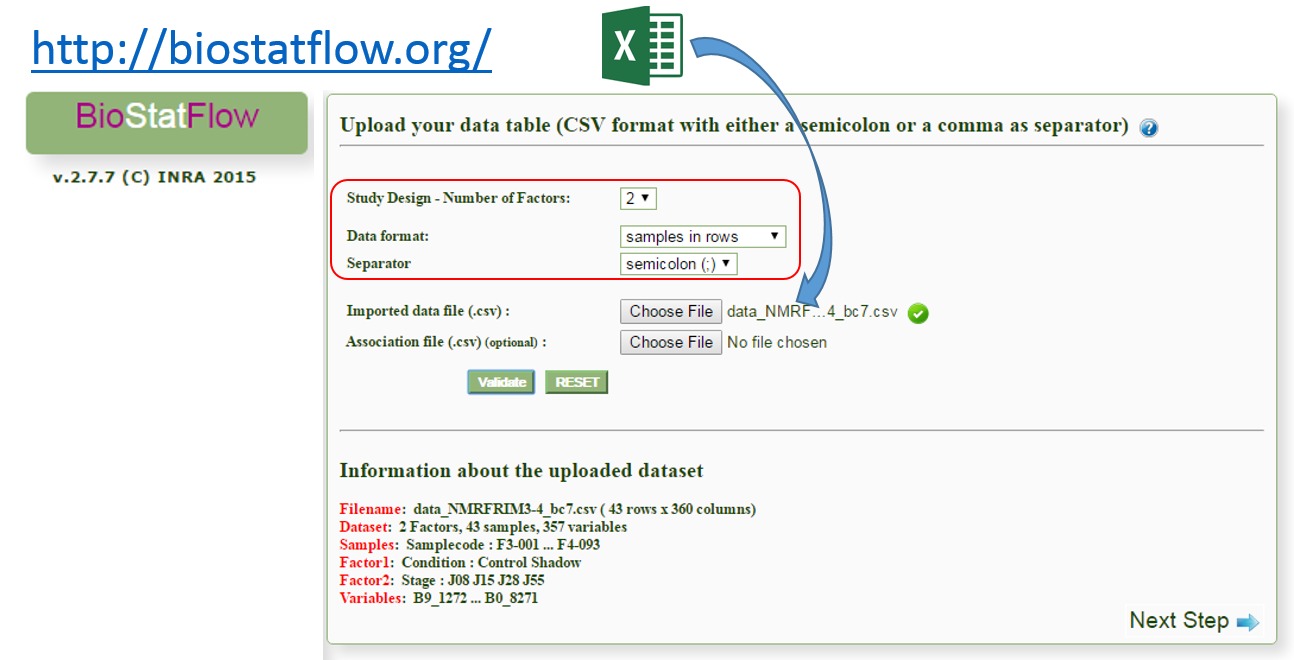
After the bucketing, you have to export the data matrix (see Export the Data matrix). The exported matrix is formatted so that we can subsequently perform statistical analysis using BioStatFlow (*) web application. Thus the data file manipulations are minimized.
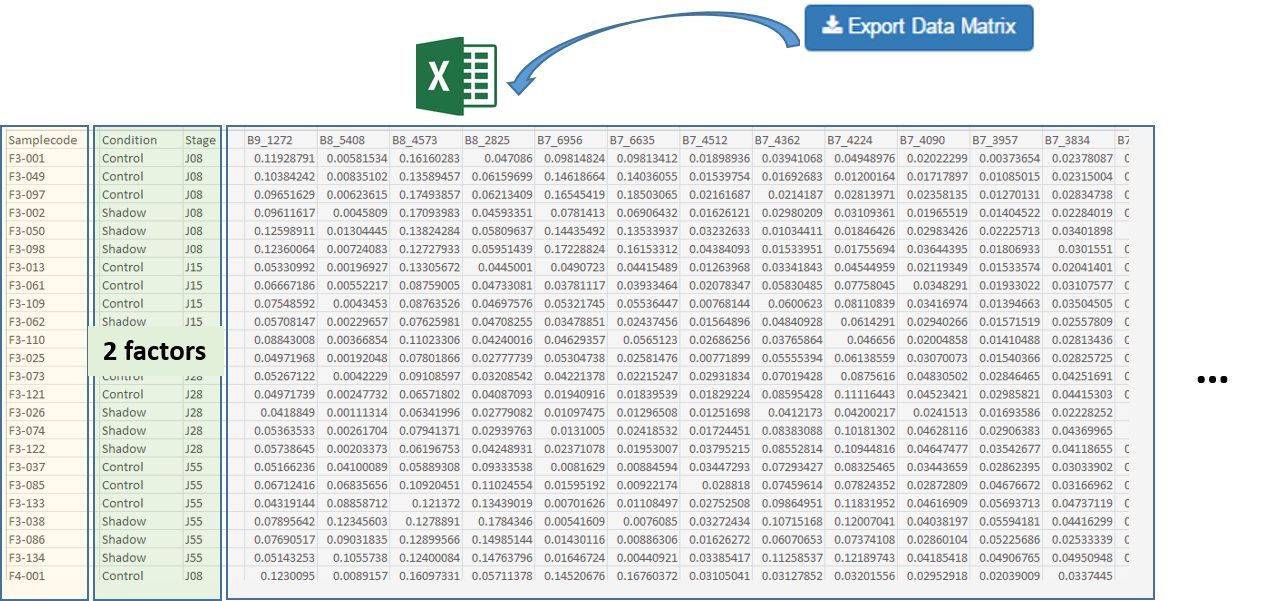
Import the Data Matrix to BioStatFlow
After exporting, the data matrix is formatted so that we can subsequently perform statistical analysis using BioStatFlow(*) web application. Thus the data file manipulations are minimized.
(*) See MetaboNews Issue 42 - February 2015
See online some slides showing a simple session of BioStatFlow in action: Example of a BioStatFlow session and the Help online of BioStatFlow
Import the Data Matrix to MetaboAnalyst
After exporting, the data matrix is formatted so that we can subsequently perform statistical analysis using MetaboAnalyst (Xia et al. 2015). See online some slides showing a simple session with MetaboAnalyst.
Xia, J., Sinelnikov, I., Han, B., and Wishart, D.S. (2015) MetaboAnalyst 3.0 - making metabolomics more meaningful . Nucl. Acids Res. 43, W251-257.
Help in the identification
Still in the embryonic stage, we currently develop tools that will greatly help in the identification. See online some slides showing about Help in the identification